Cellular motions and thermal fluctuations: the Brownian ratchet
Citation: C. S. Peskin, G. M. Odell, G. F. Oster (1993).
Link: Zotero
Summary
- Key finding:
- Method:
- Limitations:
- Notes in my words:
Relevance to my research
- Directly relevant
- Background
- Methods
Quotes / Figures
The systems we address here are different from those usually considered protein motors (e.g., myosin, dynein, kinesin), but such motors may be Brownian ratchets as well (1-4).
Parameters:
| Symbol | quantity that the symbol represents | expression |
|---|---|---|
| The dimensionless work done against the load in adding one monomer | ||
| Load force of the barrier (i.e. force that the polymer has to overcome to polymerize) | — | |
| distance between the tip of the polymer and the barrier | — |
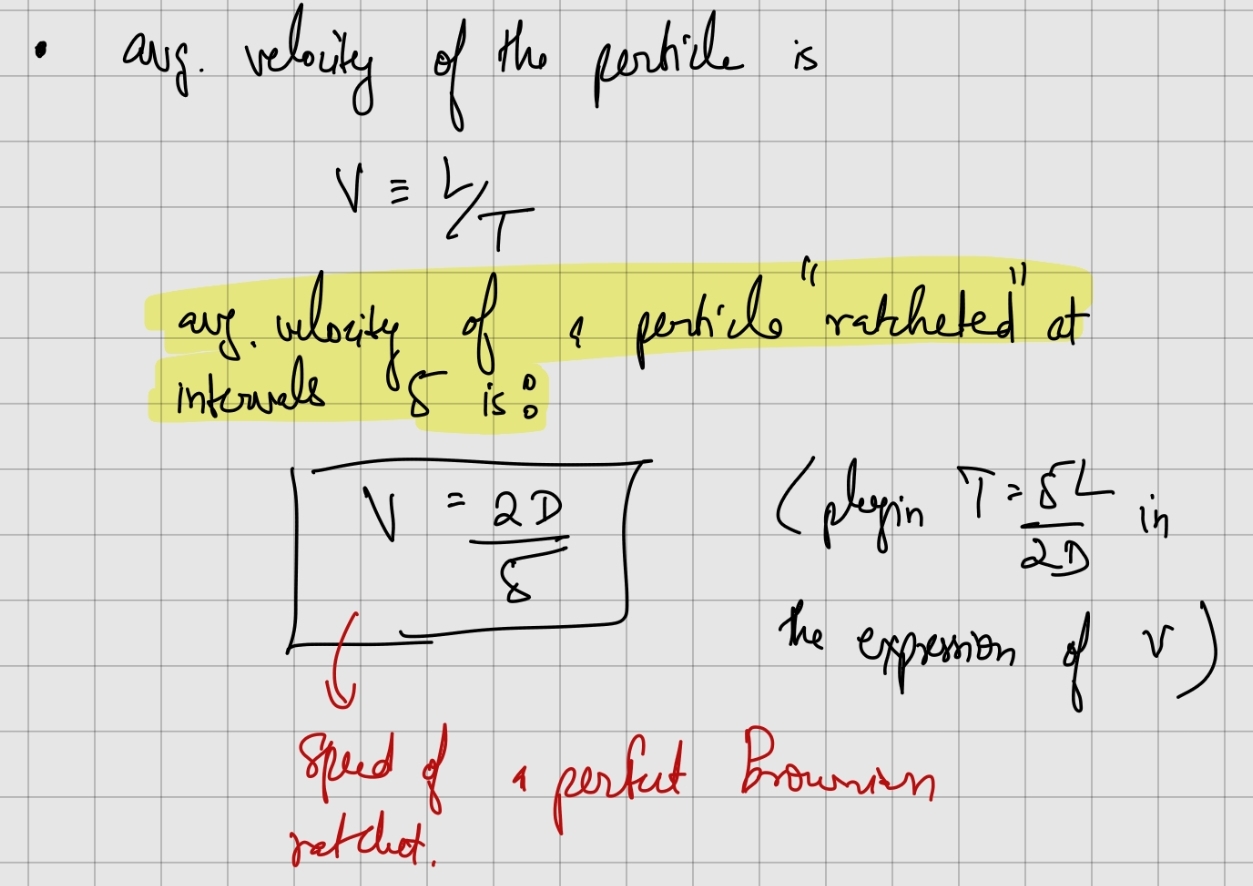
Speed of a perfect Brownian ratchet:
Note that as the ratchet interval, , decreases, the ratchet velocity increases.

If no barrier were present, actin could polymerize at a maximum velocity of ,um/s at 25 , M concentration of actin monomers
Cellular filopodia protrudes at 0.16 m /s